Data
The data used in our benchmarking study are two randomly generated DNA sequences (generated from here), each of length 1000 represented by bases C, G, A, and T. We used a MacBook Pro with an NVIDIA GPU to run the benchmarks.
Results
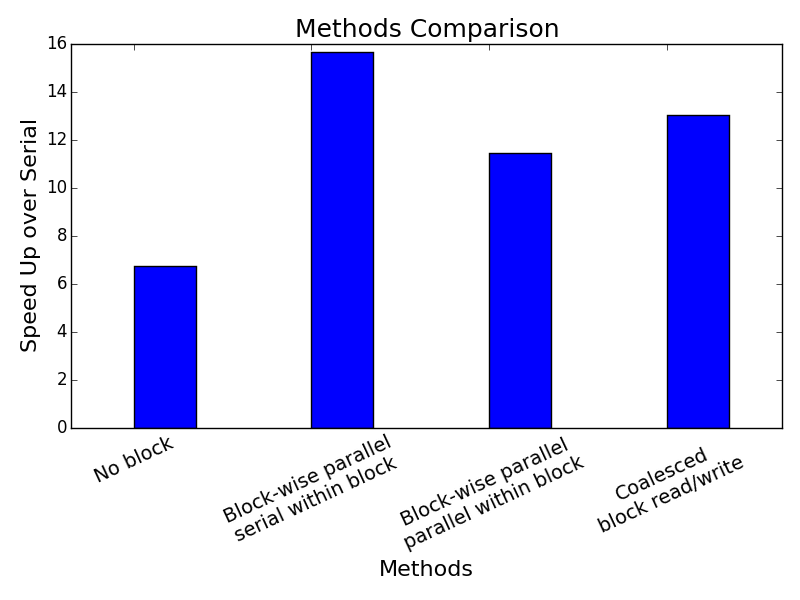
Below is a chart showing the speed-ups of the parallel algorithms using a local buffer size of 32 by 32, or 1,024 workers, over the serial dynamic programming implementation using numpy. As expected, coalesced memory access achieved higher speedup at 13 times versus non-coalesced memory access at 11.5 times. Surprisingly, block-wise parallel with serial within-block updates achieved the fastest speed up at almost 16 times. Given that we initialized the same number of workers across all parallel algorithms in this study, this outperformance could very well be hardware-dependent. We suspect that it likely stems from serialized reads when multiple threads access the same location in global memory in other parallel within block implementations.
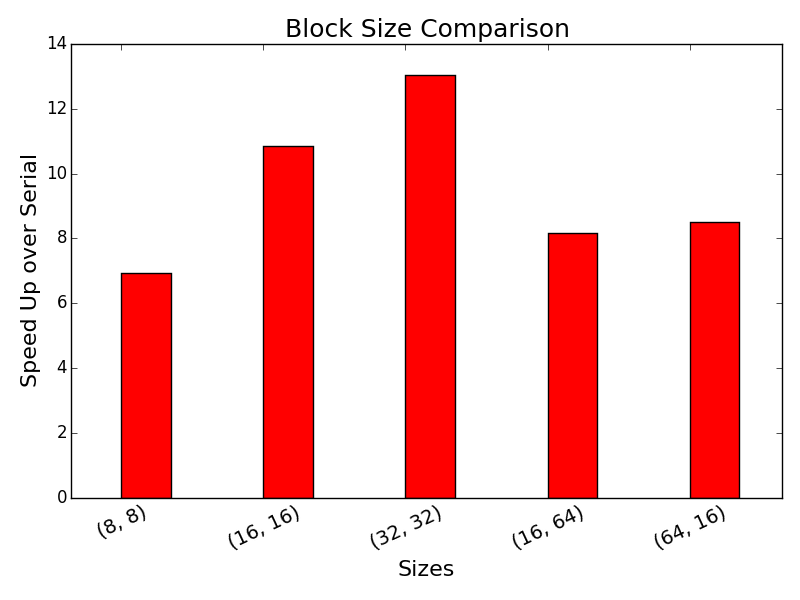
We also explored different buffer size configurations using the coelasced block read/write implememtation. As shown below, the larger the size of the workgroup, the higher the speed-up from 8 by 8 to 32 by 32. On the other hand, 32 by 32 is faster than both 16 by 64 and 64 by 16. This is likely due to the fact that the alignment matrix of our test case is a perfect square and a workgroup arranged in a square could be positioned nicely along the anti-diagonals to achive more parallelism. Finally, we found that bank conflicts are not a big issue with the parallel implementations, since it is unlikely for the threads to access the same column at the same time.